May 19, 2018 by akhilendra
Data Exploration in R with GGPLOT2 & Standard Functions
In this post, we are going to perform high level data exploration in R programming language using GGPLOT2 and its standard plotting functions. So if you are looking on information on how to make plots and graphs in R using ggplot2 or standard r functions, you will find this post useful. You can simply change the name of the data sets and other parameters to reuse them for your project.
I am not going to add details about data because the purpose is to show few common and important commands.
if you have any question, please leave your question or comment.
Minimum Steps for exploration:
- Importing the data set into R
- Understanding the structure of data set
- Graphical exploration
- Descriptive statistics
- Insights from the data set
Data Exploration in R
please make sure you set the working directory correctly so that R can find your file. please note i have included the output for reference but you will need to print it in your machine to understand it better.
I have used GGPLOT2 and standard R functions for plotting. if you want to use GGPLOT2, you will first need to install the package.
#Importing the data set into R
library(rJava)
library(xlsxjars)
library(xlsx)
lung= read.xlsx("LungCap_Dataset.xls", sheetIndex = 1, header = TRUE)
lung
Output
library(rJava) > library(xlsxjars) > library(xlsx) > lung= read.xlsx("LungCap_Dataset.xls", sheetIndex = 1, header = TRUE) > lung LungCap.cc. Age..years. Height.inches. Smoke Gender Caesarean 1 6.475 6 62.1 no male no 2 10.125 18 74.7 yes female no 3 9.550 16 69.7 no female yes 4 11.125 14 71.0 no male no 5 4.800 5 56.9 no male no 6 6.225 11 58.7 no female no 7 4.950 8 63.3 no male yes 8 7.325 11 70.4 no male no 9 8.875 15 70.5 no male no 10 6.800 11 59.2 no male no 11 11.500 19 76.4 no male yes 12 10.925 17 71.7 no male no 13 6.525 12 57.5 no male no 14 6.000 10 61.1 no female no 15 7.825 10 61.2 no male no 16 9.525 13 63.5 no male yes 17 7.875 15 59.2 no male no 18 5.050 8 56.1 no male no 19 7.025 11 61.2 yes female no 20 9.525 14 70.6 no female no 21 3.975 6 57.3 no male no 22 5.325 8 59.7 no female no 23 10.025 16 72.4 no male no 24 8.725 11 68.0 no male yes 25 9.375 11 65.7 no female no 26 8.350 12 61.3 no male yes 27 6.750 12 60.7 no female no 28 9.025 9 65.6 no male no 29 1.125 4 48.7 no female no 30 10.475 18 72.0 yes female no 31 4.650 4 53.7 no female no 32 7.725 13 64.7 no male no 33 10.600 13 69.3 no male no 34 11.025 13 65.6 no male yes 35 8.650 12 67.8 no male no 36 8.825 10 65.5 no male no 37 4.200 6 52.7 no male no 38 8.775 9 63.6 no male no 39 6.325 11 64.6 no female no 40 11.325 17 77.7 no male no 41 8.225 14 65.4 no female no 42 10.725 17 72.5 no female yes 43 5.875 8 58.9 no female no 44 7.275 12 67.7 no male no 45 1.575 6 49.3 no male no 46 6.700 11 62.6 no female no 47 7.650 11 61.7 no male yes 48 8.000 12 64.7 no female no 49 12.950 17 74.9 no male no 50 7.350 7 61.6 no male no 51 9.625 15 66.4 no male no 52 12.425 15 74.1 no male no 53 7.400 11 65.3 no male no 54 4.875 10 61.4 no male no 55 12.225 18 79.6 no male no 56 4.250 6 52.9 no male no 57 8.200 13 65.6 no male no 58 11.400 19 79.1 no male no 59 4.625 9 56.8 no female no 60 7.825 12 65.4 no male yes 61 6.700 12 57.9 no female no 62 9.200 14 68.2 no male no 63 6.950 9 61.4 no female no 64 6.850 13 58.7 no female no 65 8.450 13 65.1 no male no 66 7.350 13 67.5 yes female no 67 5.375 11 59.3 yes female no 68 7.375 11 63.0 no female no 69 8.600 11 64.4 no female no 70 7.900 12 68.0 no male no 71 8.500 14 61.4 no female yes 72 9.700 11 72.4 no male no 73 5.125 11 51.5 no female yes 74 7.825 13 71.0 no female no 75 6.250 13 61.8 no female no 76 4.975 12 62.6 yes female yes 77 7.500 14 66.6 no male no 78 5.875 9 59.0 no female no 79 10.050 17 70.4 no female no 80 10.800 11 69.8 no male no 81 7.350 12 63.0 no female no 82 11.900 16 69.3 no male no 83 12.050 17 72.2 no male no 84 11.575 19 78.2 no female no 85 6.200 14 61.1 no female no 86 6.125 12 63.3 no female yes 87 13.875 19 78.4 no male yes 88 7.750 11 63.5 no female no 89 7.475 15 63.0 yes female no 90 11.575 19 75.5 no male no 91 6.950 9 63.9 no male yes 92 9.200 14 69.4 no male no 93 9.750 13 72.8 yes male no 94 9.650 14 65.2 no female no 95 11.750 19 78.0 yes female yes 96 10.825 18 75.7 no female no 97 7.550 16 71.1 yes male no 98 6.950 7 64.7 no male no 99 10.675 16 74.9 no male no 100 6.100 10 57.0 no male no 101 8.025 13 66.2 yes male no 102 9.225 14 66.9 no male no 103 3.450 13 58.5 no female yes 104 10.725 16 75.6 no female no 105 7.950 16 67.3 no female no 106 3.425 5 51.7 no female no 107 10.875 16 75.5 no male no 108 8.625 12 64.8 no male no 109 6.450 7 63.2 no male no 110 3.100 7 52.1 no male no 111 10.425 15 70.6 no male no 112 12.150 18 76.3 no female no 113 1.850 8 49.8 no female no 114 5.875 3 55.9 no male no 115 9.125 15 73.4 no male no 116 8.975 15 67.5 no female yes 117 3.750 7 50.3 no female no 118 10.275 18 71.0 no male no 119 6.675 8 54.9 no female no 120 11.775 17 76.9 yes male no 121 8.550 16 67.9 no male no 122 6.450 12 61.0 yes male yes 123 13.200 17 78.6 no male yes 124 11.550 16 75.7 no male no 125 12.950 19 79.6 no male yes 126 7.825 12 67.5 no female yes 127 10.550 17 71.8 no male yes 128 11.700 19 76.2 no female yes 129 3.650 12 56.6 no male yes 130 6.650 12 60.0 yes female yes 131 10.425 15 67.2 no male no 132 12.925 17 75.7 no female no 133 7.450 13 61.1 no male no 134 8.600 12 60.1 no female no 135 10.650 16 74.4 no male no 136 4.725 13 65.5 no female no 137 7.550 15 69.3 no female no 138 10.175 15 71.4 no female no 139 6.450 14 61.4 no male no 140 9.475 13 67.4 no male no 141 4.975 6 58.4 no male no 142 9.900 18 70.9 no female no 143 10.200 18 68.6 no female no 144 12.400 18 81.8 no male no 145 6.850 9 65.7 no female yes 146 11.825 17 73.9 no female no 147 8.625 14 66.8 no female yes 148 11.350 14 70.5 no male no 149 8.225 14 64.0 no female no 150 0.507 3 51.6 no female yes 151 5.075 11 61.2 no male no 152 6.450 8 62.7 no male no 153 6.725 9 56.1 no male no 154 4.525 8 55.5 no female no 155 9.275 16 67.2 no female no 156 2.850 7 51.4 no female no 157 9.350 11 71.2 yes male no 158 5.550 5 55.8 no female yes 159 10.350 16 73.5 no male yes 160 6.625 11 62.4 no male yes 161 9.725 16 68.6 no female yes 162 4.900 10 56.8 no female no 163 10.475 12 69.7 no male no 164 10.850 19 70.9 no male no 165 5.150 7 58.4 no female no 166 4.425 8 56.6 no male no [ reached getOption("max.print") -- omitted 559 rows ]
Summarizing Data in R for exploration
let’s summarize data and go through some of the most common parameters.
# summaries data to understand it further summary(lung) #use str to understand the structure of dataframe str(lung) # use table to get the frequency table(lung$Age..years.) # get mean & standard deviation mean(lung$Age..years.) sd(lung$Age..years.)
Output
> summary(lung) LungCap.cc. Age..years. Height.inches. Smoke Gender Caesarean Min. : 0.507 Min. : 3.00 Min. :45.30 no :648 female:358 no :561 1st Qu.: 6.150 1st Qu.: 9.00 1st Qu.:59.90 yes: 77 male :367 yes:164 Median : 8.000 Median :13.00 Median :65.40 Mean : 7.863 Mean :12.33 Mean :64.84 3rd Qu.: 9.800 3rd Qu.:15.00 3rd Qu.:70.30 Max. :14.675 Max. :19.00 Max. :81.80
> str(lung) 'data.frame': 725 obs. of 6 variables: $ LungCap.cc. : num 6.47 10.12 9.55 11.12 4.8 ... $ Age..years. : num 6 18 16 14 5 11 8 11 15 11 ... $ Height.inches.: num 62.1 74.7 69.7 71 56.9 58.7 63.3 70.4 70.5 59.2 ... $ Smoke : Factor w/ 2 levels "no","yes": 1 2 1 1 1 1 1 1 1 1 ... $ Gender : Factor w/ 2 levels "female","male": 2 1 1 2 2 1 2 2 2 2 ... $ Caesarean : Factor w/ 2 levels "no","yes": 1 1 2 1 1 1 2 1 1 1 ... > table(lung$Age..years.) 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 13 6 20 25 37 41 40 51 58 68 69 56 64 54 43 43 37
> mean(lung$Age..years.) [1] 12.3269 > sd(lung$Age..years.) [1] 4.00475
Random sampling
sample(lung)
random sampling with size
sample(lung, 5)
Create histograms in R with standard hist function
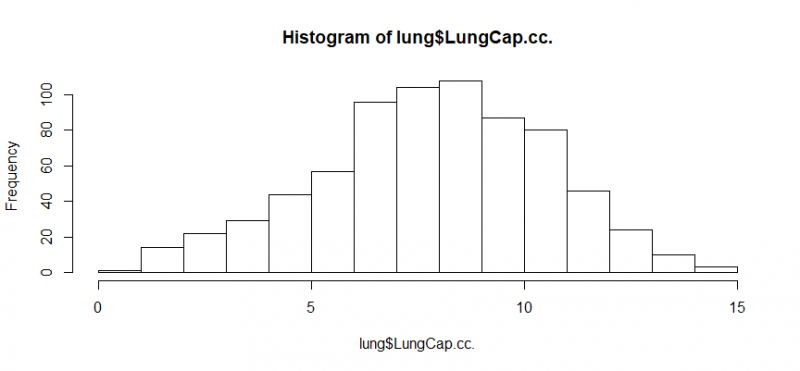
hist(lung$LungCap.cc.)
hist(lung$Age..years.)
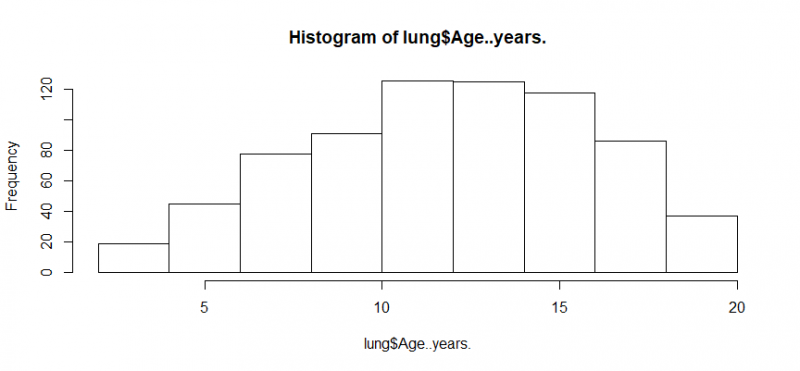
stem(lung$LungCap.cc.,scale = 1,width = 80,atom = 1e-08)
0 | 5 1 | 012356678999 2 | 00033345666777778999999 3 | 0011222344445667777788999999 4 | 001122233333344445555556666677777788899999 5 | 00000001111122222223333334444455566666677777778899999999 6 | 00000111111111111122222222222223333334445555555555555566666666667777+13 7 | 00000000001111112222233333333333344444444444444555555556666666666677+20 8 | 00000000000000001111111122222222333333333444444444444445555555556666+32 9 | 00000000000011111111122222222223333333444445555555555666666667777777+11 10 | 00000000111111112222222333333444444444555555555566666667777777777788 11 | 0000001111111222222333344455556666677777888889999 12 | 01111222223333444456799 13 | 000111234449 14 | 467
stem(lung$Height.inches.,scale = 1,width = 80,atom = 1e-08)
The decimal point is at the | 44 | 3 46 | 604478 48 | 01278902389 50 | 357000124556777999 52 | 001777888990223567778899 54 | 2357899901111244555566666677899 56 | 001112334556666667788889901123334445789 58 | 000334444455555677778889990012222223333444457777889999 60 | 0000111222233444445566677789001111122233444444555566667788889999 62 | 00001111112344455666666778888990000122233333334444445555566667779999 64 | 00011111233444455667777778889990000111123333344444444555555566666677 66 | 00000011122233333444445555666678899990112223333444455555556666777778 68 | 00001111222223334444566666677788889990000111222333333344444466777788 70 | 0011222344445555668899999000011112222233444455556667788899999 72 | 00012223444455555667889900111233344555555566667788999 74 | 000012222334456677899901223445556677778889 76 | 012233456688992467 78 | 024469913668 80 | 388
Plot Functions in R
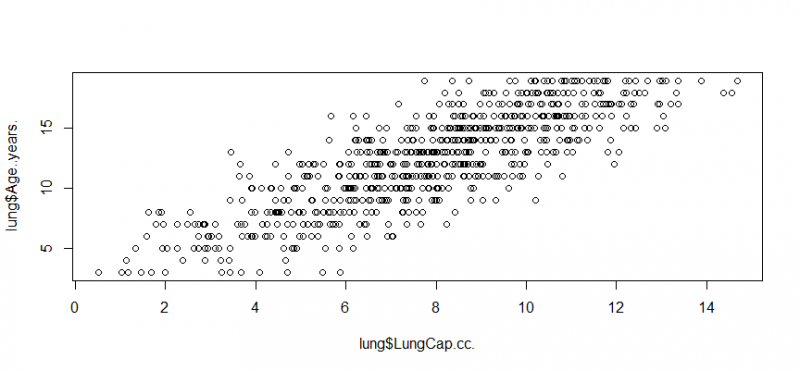
plot(lung$LungCap.cc.,lung$Age..years.)
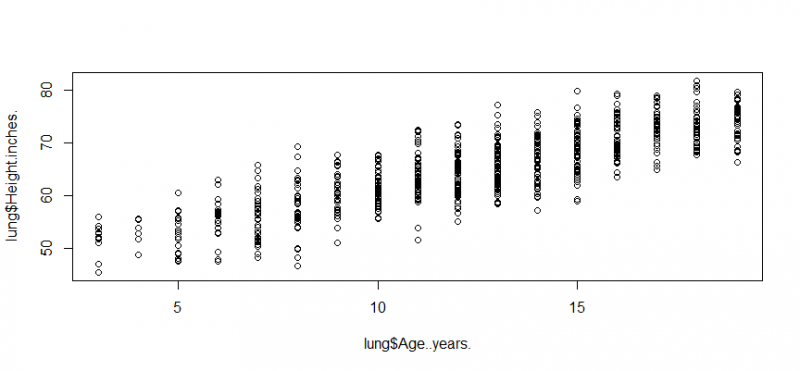
plot(lung$Age..years.,lung$Height.inches.)
Plotting with GGPLOT2
if you need details about ggplot2 library, please visit my previous post on it. Click Here.
now let’s use ggplot2
library(ggplot2) # age..years & lung$Height.inches. are x & y axis here # i have used shape to group them based on gender qplot(lung$Age..years.,lung$Height.inches.,data = lung,shape=as.factor(Gender))
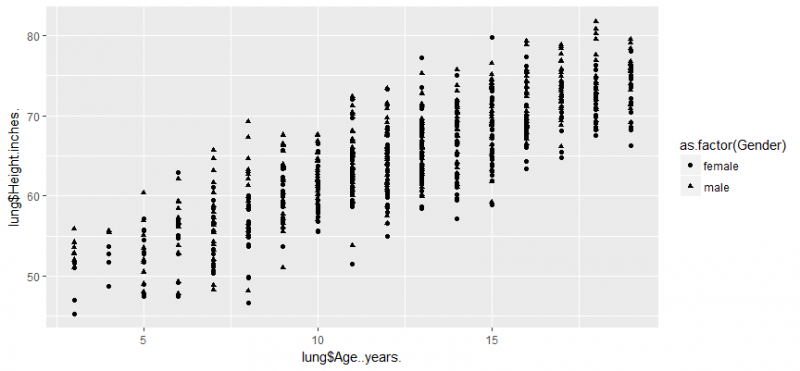
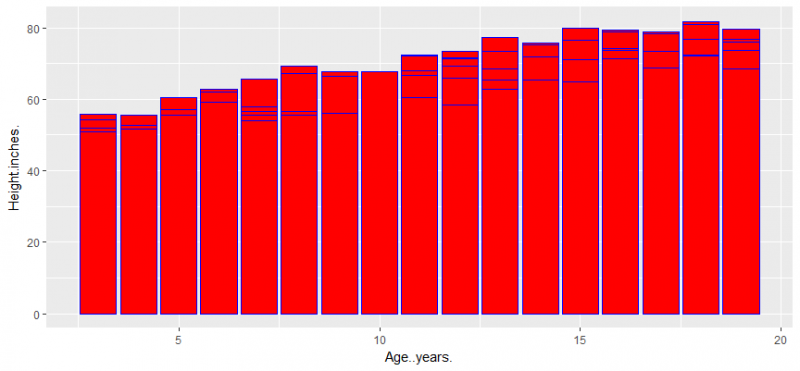
let’s create a bar chart and add color by using fill and colour
ggplot(data = lung, aes(x=Age..years., y=Height.inches.)) + geom_bar(stat = "identity",position = "dodge",colour= "blue", fill = "red")
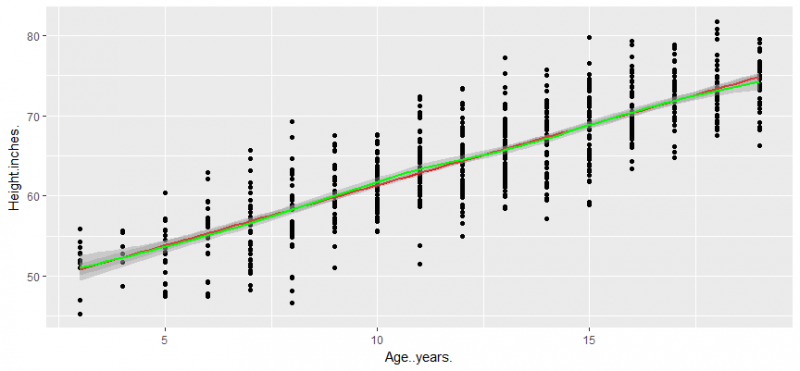
now create a scattered plot with ggplot2 with a fitted line
sctplot<-ggplot(data = lung, aes(x=Age..years., y=Height.inches.)) + geom_point() sctplot<- sctplot + geom_smooth(method ="lm", col="red") sctplot + geom_smooth(col="green")
let’s also add boxplot in the mix
boxplot(lung,las=1)
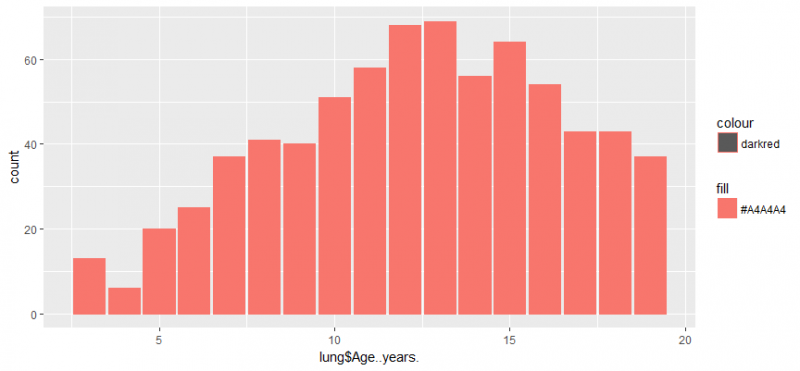
Let’s create more charts using already used commands in this post:
g <- ggplot(lung, aes(lung$Age..years.)) g + geom_bar() g + geom_bar(aes(weight = lung$Age..years.)) g + geom_bar(aes(fill = '#A4A4A4',, color="darkred"))
As mentioned earlier, objective of this post is to show some of these useful R and ggplot2 commands. If you are looking for something specific, please share your question, i will try to answer that.

Leave a Reply